Research History
Budding yeast centromere DNA and kinetochore proteins.
The budding yeast centromere is an excellent model system for investigating the molecular mechanisms involved in chromosome segregation and genetic inheritance. For many years my research group has studied the S. cerevisiae centromere, initially focusing on the CEN DNA, and later working to identify the proteins associated with the CEN DNA that assembles the kinetochore structure. In the early 1990's using a genetic assay based on a mutant CEN DNA, we identified a remarkable new protein, Cse4p, which is required to assemble centromere-specific nucleosomes and for accurate chromosome segregation during mitosis (Gaudet, A. and M. Fitzgerald-Hayes 1987, Gaudet, A. and M. Fitzgerald-Hayes 1989). The Cse4p protein masquerades as an H3 histone protein and becomes incorporated into a nucleosome in place of H3. The specialized nucleosome with Cse4p forms only at the centromere (Stoler, S. et al. 1995, Meluh, P.B. et. al 1998). To further investigate how the Cse4p protein accomplishes this task, we plan to use genetic, biochemical and molecular approaches to study the targeting mechanism that localizes the Cse4p-nucleosome to the yeast centromere.
Yeast centromeres are relevant to understanding human centromeres.
The budding yeast centromere DNA (CEN), and many centromere proteins have now been characterized (Henikoff, S. et al. 2000, Ahmad, K. and S. Henikoff 2002). Each budding yeast centromere has a single microtubule (MT) spindle fiber attachment site. Compared to the centromeres in other eukaryotic organisms the yeast centromere has a relatively simple structure with only 125 bp of CEN DNA (Fitzgerald-Hayes, M. et al. 1982, Clarke, L. and J. Carbon 1983, Ng, R., J. Ness, and J. Carbon 1986). To date, more than 35 yeast centromere proteins have been identified and work is underway to determine how these proteins build a functional kinetochore structure at the centromere (Cheeseman, I.M. et al. 2002). Learning about how the budding yeast centromere works is relevant to a better understanding of human centromere structure and function. Cse4p has a very similar ortholog at human centromeres called CENP-A (Sullivan, K.F. et al. 1994). Other yeast centromere proteins also have potential orthologs in humans such as CENP-B and CENP-C, which resemble the yeast Cbf1p and Mif2p proteins, respectively (Shelby, R.D. et al. 1997, Meluh, P.B. and D. Koshland 1995, Iwahara, J. et al. 1998). The significance of understanding these biological processes in yeast and humans has wide implications. Aberrant centromere function causes chromosome missegregation and altered genetic inheritance, often with drastic consequences for the organism. Genome instability and chromosome aneuploidy can result from faulty centromere function and are common features of most cancer cells and many birth disorders.
Cse4p is a member of a new CenH3 protein family.
Cse4p and CENP-A were the first members of the CenH3 (centromere-specific H3-like) family of proteins to be characterized, including Drosophila (Cid), Arabidopsis (HTR12), and C. elegans (HCP-4) (Vermaak, D. et al. 2002, Talbert, P.B. et al. 2002, Buchwitz, B.J. et al. 1999. The CenH3 proteins all contain histone fold domains (HFD) with strong amino acid homology with the HFD of the core histone, H3. The CenH3 proteins also contain N-termini of different lengths, which lack amino acid homology. The kinetochore structure varies in different organisms, which have different DNA sequences and protein composition (except for the very highly conserved histone proteins). The fact that Cse4p belongs to a family of conserved proteins common to very diverse kinetochore structures is very intriguing. It strongly supports our contention that what we learn about Cse4p is relevant to our understanding of mammalian kinetochores. Interestingly, preliminary results indicate that the Cse4p sequences necessary for targeting to the CEN in yeast overlap with the regions of CENP-A and Cid that are required to localize to the human and fly centromeres, respectively. We plan to continue to use Cse4p and the budding yeast system to unravel the pathways necessary to assemble specialized nucleosomes at the centromere.
The budding yeast advantage for centromere studies.
Studying Cse4p function in budding yeast has many experimental advantages. S. cerevisiae cells are very amenable to genetic approaches and recombinant DNA methods as well as to biochemical fractionation and microscopy techniques. The S. cerevisiae genome sequence is complete and many genes have been located on the yeast genome map. In addition, the yeast cells divide very rapidly and execute a well-characterized cell division cycle, both important features for studying chromosome movement. Yeast cells adopt specific cell morphologies that reflect the cell cycle stage of the cell. For example, when the temperature sensitive cse4-1 cells are shifted from the permissive to the non-permissive growth temperature, the cse4-1 cells arrest with a large budded morphology indicative of cells in mitosis.
Budding yeast CEN DNA and specialized chromatin.
The entire S. cerevisiae CEN DNA locus encompasses only ~125 bp of DNA, and contains three centromere DNA elements: CDEI (8 bp), CDEII (~80 bp) and CDEIII (26 bp) conserved among the 16 budding yeast centromeres (Fleig, U. et al. 1995) (Figure 1). The yeast centromere proteins interact with the CEN DNA elements to assemble chromatin and construct the kinetochore. The Cbf1p helix turn helix protein binds to CDEI DNA as a homo-dimer (CBF1/CDEI) (Niedenthal, R. et al. 1993). All 16 CEN CDEII DNA regions are extremely A+T-rich (>90%) and each contains an intrinsic DNA bend. The conserved length of ~80 bp for CDEII is also the length of the DNA needed to wrap the helix once around an octamer core of histone proteins. We showed by mutagenesis studies that the length, base composition and secondary structure (bent or unbent) are the primary physical characteristics of CDEII DNA that contribute to centromere function and centromere chromatin assembly (Gaudet, A. and M. Fitzgerald-Hayes 1987, Gaudet, A. and M. Fitzgerald-Hayes 1989, Murphy, M.R. et al. 1991). The specialized chromatin at the centromere acts as a foundation for the assembly of a kinetochore-spindle fiber attachment site. Kinetochore assembly requires the CDEIII CEN DNA sequence, and the multi-protein CBF3 complex (CFB3/CDEIII). We showed that changing a single base pair in the CDEIII DNA sequence abolishes centromere function, destroys the protein complexes at the centromere and is lethal to the yeast cells (McGrew, J. et al. 1986).
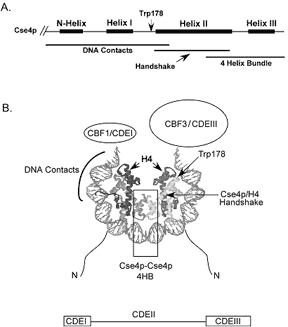
Figure 1. Cse4p/CEN specific nucleosome.
(A) Cse4p HFD with helical domains. (B) Cse4p-tetramer binds CEN DNA;
recruited to CEN by CBF3/CDEIII.
Gaudet, A. and M. Fitzgerald-Hayes, Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Molecular and Cellular Biology, 1987. 7: p. 68-75.
Gaudet, A. and M. Fitzgerald-Hayes, Mutations in CEN3 cause aberrant chromosome segregation during meiosis in Saccharomyces cerevisiae. Geneitcs, 1989. 121: p. 447-489.
Stoler, S., K.C. Keith, K.E. Curnick, and M. Fitzgerald-Hayes, A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes and Development, 1995. 9: p. 573-586.
Meluh, P.B., P. Yang, L. Glowczewski, D. Koshland, and M.M. Smith, Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell, 1998. 94: p. 607-613.
Henikoff, S., K. Ahmad, J.S. Platero, and B. van Steensel, Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A, 2000. 97(2): p. 716-721.
Ahmad, K. and S. Henikoff, Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A, 2002. 99 Suppl 4: p. 16477-84.
Fitzgerald-Hayes, M., L. Clarke, and J. Carbon, Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell, 1982. 29: p. 235-244.
Clarke, L. and J. Carbon, Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature, 1983. 305: p. 23-28.
Ng, R., J. Ness, and J. Carbon, Structural studies on centromeres in the yeast Saccharomyces cerevisiae, in Extrachromosomal Elements in Lower Eukaryotes, A.H. R. B. Wickner, A. M. Lambowitz, I. C. Gunsalus, and A. Hollaender, Editor. 1986, Plenum Publishing Corp. p. 479-492.
Cheeseman, I.M., D.G. Drubin, and G. Barnes, Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol, 2002. 157(2): p. 199-203.
Sullivan, K.F., M. Hechenberger, and K. Masri, Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. Journal of Cell Biology, 1994. 127: p. 581-592.
Shelby, R.D., O. Vafa, and K.F. Sullivan, Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. Journal of Cell Biology, 1997. 136: p. 501-513.
Meluh, P.B. and D. Koshland, Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein, CENP-C. Molecular Biology of the Cell, 1995. 6: p. 793-807.
Iwahara, J., T. Kigawa, K. Kitagawa, H. Masumoto, T. Okazaki, and S.
Yokoyama, A helix-turn-helix structure unit in human centromere protein
B (CENP-B). EMBO, 1998. 17: p. 827-837.
Vermaak, D., H.S. Hayden, and S. Henikoff, Centromere targeting element
within the histone fold domain of Cid. Mol Cell Biol, 2002. 22(21): p.
7553-61.
Talbert, P.B., R. Masuelli, A.P. Tyagi, L. Comai, and S. Henikoff, Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell, 2002. 14(5): p. 1053-66.
Buchwitz, B.J., K. Ahmad, L.L. Moore, M.B. Roth, and S. Henikoff, A histone-H3-like protein in C. elegans. Nature, 1999. 401(6753): p. 547-8.
Fleig, U., J.D. Beinhauer, and J.H. Hegemann, Functional selection for the centromere DNA from yeast chromosome VII. Nucleic Acids Research, 1995. 23: p. 922-924.
Niedenthal, R., K., M. Sen-Gupta, A. Wilmen, and J.H. Hegemann, Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acid Research, 1993. 21: p. 4726-4733.
Murphy, M.R., D.M. Fowlkes, and M. Fitzgerald-Hayes, Analysis of centromere function in Saccharomyces cerevisiae using synthetic centromere mutants. Chromosoma, 1991. 101: p. 189-197.
McGrew, J., B. Diehl, and M. Fitzgerald-Hayes, Single base-pair mutations
in centromere element III cause aberrant chromosome segregation in Saccharomyces
cerevisiae. Molecular and Cellular Biology, 1986. 6: p. 530-538.